CompLearn Example: Evolution at a Glance
The following example uses gene sequences of 34 mammals, which we have also included with the software.
Step 1: Create a Distance Matrix
$ ncd -b -d examples/34-mammals examples/34-mammals
This creates a square distance matrix using the gene sequences in the examples/34-mammals directory of the CompLearn distribution. With the -b option, a binary file called distmatrix.clb will be created by default. The results of this command are also output to stdout.
Step 2: Create a Tree
$ maketree distmatrix.clb
Here, we use the previously created distance matrix to create an unrooted binary tree. The resulting tree is output as a .dot file called treefile.dot by default. The contents of the resulting .dot file describe how the nodes of the tree are connected and how they are labeled. Please note: because we are generating a tree from a rather large matrix (34x34), maketree may take several hours to complete. However, feel free to take a peek at treefile.dot while it is still in progress.
Step 3: Lay Out Your Tree
$ neato -Tps -Gsize=7,7 treefile.dot > 34mammals-unrooted.ps
This neato command will create a visual representation of your tree in postscript format. This particular postscript file is laid out on a 7x7 inch drawing area.
If instead you prefer a browser-compatible format, such as .png you can use the following command.
$ neato -Tpng treefile.dot > 34mammals-unrooted.png
neato supports other popular formats such as .jpg and .gif.
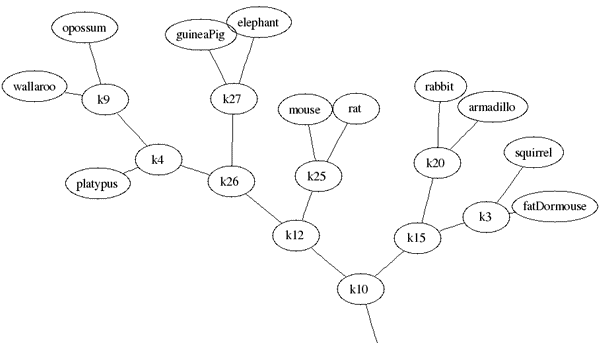
Step 4: Analyze Your Results
As you can see in the resulting tree layout (click here for complete tree), "like" mammals are clustered together as you would expect. Of particular note, notice primates occupy their own major branch of the tree, and, as seen in the portion below, marsupials are grouped together. The placement of "elephant" in the midst of rodents, however, is quite curious and requires further investigation. How do you think an elephant likes being in the middle of so many mice?